BioTechniques News
Beatrice Bowlby

Researchers at Duke University (NC, USA) have developed CellREADR, a new piece of RNA-based editing technology that operates by targeting cells instead of genes.
CellREADR is a string of RNA comprised of three distinct sections: a sensor, a stop sign and a set of blueprints [1]. The sections are customizable and should be able to facilitate a range of objectives due to CellREADR’s ability to add any protein to a user-specified cell type. The authors have high hopes for its potential to progress the treatment of complex diseases, such as drug-resistant epilepsy.
CellREADR’s monitoring and control systems rely on a specific enzyme – the ADAR enzyme – to operate. The ADAR enzyme is responsible for responding to the presence of double-stranded RNA, which occurs almost exclusively in relation to viruses and viral replication [2]. Given ADAR’s central role in the workings of this RNA-based editing tool, it’s a good thing, then, that it is present in every animal cell.
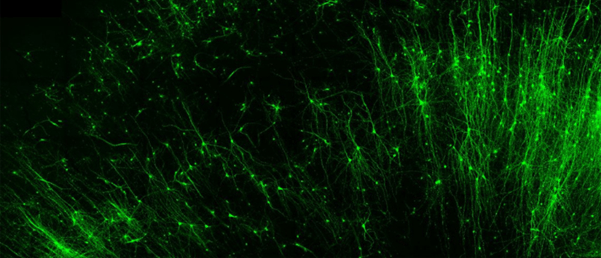
So far, the new platform has demonstrated compatibility with both rodent and human brain tissue. Because ADAR exists in all animal cells, the researchers expect that the technology will be relatively simple to adapt to other creatures besides rodents and humans.
 Novel CRISPR-Cas9 platform delivers gene editing across cell membranes
Novel CRISPR-Cas9 platform delivers gene editing across cell membranes
A new class of spherical nucleic acids has been developed to deliver CRISPR-Cas9 machinery across cell membranes and into the nucleus, without the need for transfection agents.
In the current study, the researchers used an RNA-based probe to introduce fluorescent tags into specific types of brain tissue, effectively labeling them. From there, they were able to activate or shut down neurons of their choosing using a light-sensitive switch. Finally, they demonstrated the capability to leverage a self-destruct enzyme to erase specific cells.
“We could actually modify specific types of cell function to manage diseases, regardless of their initial genetic predisposition,” senior author Josh Huang reported. “That’s not possible with current therapies or medicine.”
They were able to do all of this by first selecting a specific cell type they wished to target and identifying the relevant RNA produced by that cell type. As each cell type produces its own signature RNA, differentiated from other cell types, they could be sure that they were targeting the exact cell type that they wanted.
They then designed a sensor sequence to compliment the target RNA strand. The sensor enters the cell and connects with the target RNA to create an artificial piece of double-stranded RNA.
Having tricked the body into thinking that there is a piece of double-stranded RNA floating around inside a cell, the ADAR enzyme is triggered and inspects the new virus-looking RNA. Once it has inspected the problem RNA, the ADAR will change a single nucleotide in that string of RNA code. By removing this specially designed nucleotide, the stop sign, the blueprints can then be read by cellular machinery to produce the desired protein.
The researchers wanted to provide scientists with the ability to alter cells and their functions at a fundamental level with the hope that this technology will one day be a disease therapy. CellREADR may be a project just taking its first steps, but the potential implications of this development are broad.
First author Yongjun Qian commented: “When I majored in pharmacology as an undergraduate, I was very naïve. I thought you could do a lot of things, like cure cancer, but actually it’s very difficult. However, now I think, yes maybe we can do it.”
The post CellREADR targets cells, not genes, for RNA-based editing across species appeared first on BioTechniques.
Full BioTechniques Article here
Powered by WPeMatico
